Designing the FragmentedMolecule Class
The point of this page is to document the design decisions which went into
the FragmentedMolecule class.
Why Do We Need a FragmentedMolecule Class?
The need for the FragmentedMolecule class was motivated by the
chemical system hierarchy consideration of
Designing the Fragmenting Component. In short, each layer of the
ChemicalSystem class will require a corresponding container for holding
fragments. This is because each layer of the ChemicalSystem introduces new
state.
FragmentedMolecule Considerations
- molecule state
The
FragmentedMoleculeclass must hold the state for a series ofMoleculeobjects. In addition to the nuclei in each fragment, this includes the number of electrons in each fragment as well as the multiplicity/spin of each fragment.
- molecule compatible
The fragments should be usable anywhere
Moleculeobjects are.
FragmentedMolecule APIs
To construct a FragmentedMolecule:
// It's assumed users already have a Molecule object
Molecule a_molecule = make_a_molecule_object();
FragmentedMolecule null; // See FragmentedBase for more details
FragmentedMolecule empty(a_molecule); // Empty
// If you already have fragmented nuclei:
FragmentedNuclei frags = make_fragments();
FragmentedMolecule has_frags(frags, a_molecule); // dispatches to next ctor
FragmentedMolecule has_frags2(frags, a_molecule.charge(), a_molecule.multiplicity());
// If you also want to set the charges and multiplicities
// Here we have fragment which is a neutral singlet and another which is a
// doublet with a charge of +1
auto charges = {0, 1};
auto multiplicities = {1, 2};
FragmentedMolecule has_frags3(frags, a_molecule, charges, multiplicities);
A couple of notes on the above:
make_a_molecule_objectandmake_fragmentsare opaque functions which respectively encapsulate creating aMoleculeand aFragmentedNucleiobject. The details of these functions are irrelevant for our purposes.Similar to
FragmentedNucleiwe expect thatFragmentedMoleculeobjects will be created as part of an algorithm rather than defining the entire state initially.Charges and multiplicities are assumed to be such that the 0-th fragment in
fragsis the neutral singlet and the 1-st is the +1 doublet.
Following from the second note we expect the following to be the usual workflow:
for(const auto& frag_i : has_frags){
auto charge, mult = compute_electronic_state(frag_i.nuclei());
frag_i.set_charge(charge);
frag_i.set_multiplicity(mult);
}
Notably this just builds off of the functionality of MoleculeView.
FragmentView Design
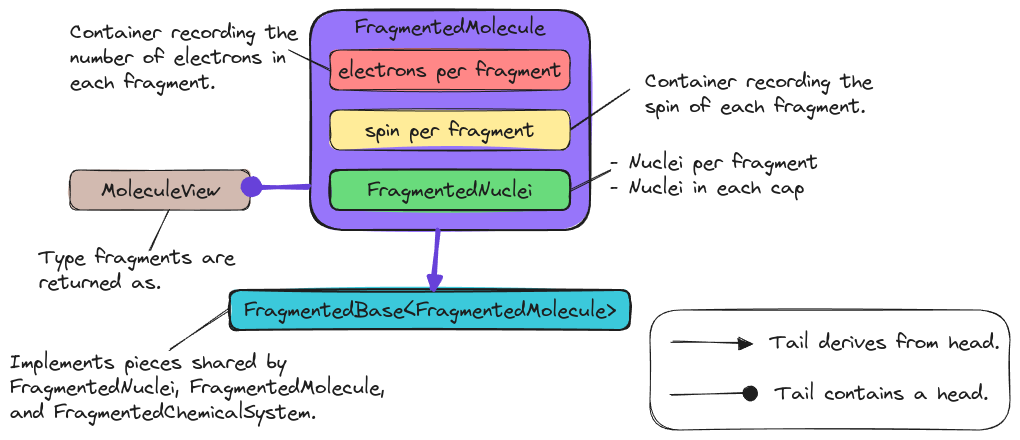
Fig. 13 State of the FragmentedMolecule class and its relationship to related
classes.
Fig. 13 shows the design of the
FragmentedMolecule class and its relationship to other classes in Chemist.
Stemming from the molecule state consideration, the
FragmentedMolecule class contains a FragmentedNuclei object and two
containers which hold the number of electrons per fragment and the
spin/multiplicity of the fragments.
Summary
- molecule state
FragmentedMoleculeobjects contain enough data to constructMoleculeViewobjects for each fragment.- molecule compatible
Fragments are returned as
MoleculeViewobjects, which can be implicitly converted toMoleculeobjects.